前言
随机样本一致性 (RANSAC) 是一种迭代方法,用于从一组包含异常值的观察数据中估计数学模型的参数,此时异常值不会对估计值产生影响。简言之,RANSAC是一种滤除异常值的常用算法。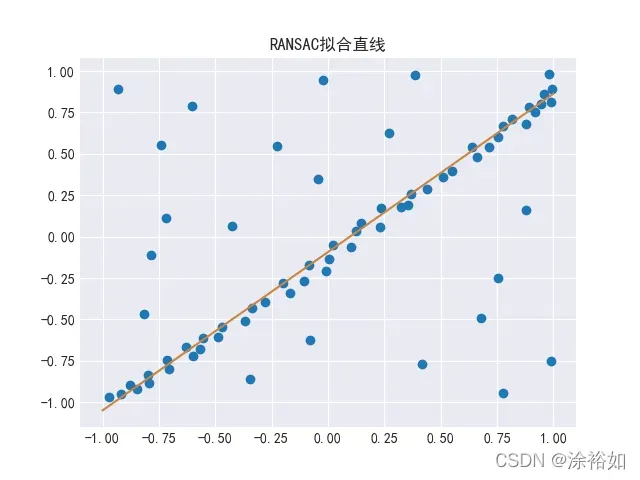
算法流程
以直线拟合为例,通用RANSAC算法的流程如下
输入:
data – 观测结果.
model – 数学模型.
n – 估计模型所需的最小样本.
k – 最大迭代次数.
t – 用于确定模型适合的数据点的阈值.
输出:
bestFit – 模型
iterations = 0
bestFit = null
bestErr = something really large
while iterations < k do
maybeInliers := n randomly selected values from data
maybeModel := model parameters fitted to maybeInliers
alsoInliers := empty set
for every point in data not in maybeInliers do
if point fits maybeModel with an error smaller than t
add point to alsoInliers
end if
end for
if the number of elements in alsoInliers is > d then
betterModel := model parameters fitted to all points in maybeInliers and alsoInliers
thisErr := a measure of how well betterModel fits these points
if thisErr < bestErr then
bestFit := betterModel
bestErr := thisErr
end if
end if
increment iterations
end while
return bestFit
Python代码
"""_summary_
RANSAC直线拟合
"""
from copy import copy
import numpy as np
from numpy.random import default_rng
rng = default_rng()
class RANSAC:
def __init__(self, n=10, k=100, t=0.05, d=10, model=None, loss=None, metric=None):
self.n = n
self.k = k
self.t = t
self.d = d
self.model = model
self.loss = loss
self.metric = metric
self.best_fit = None
self.best_error = np.inf
def fit(self, X, y):
for _ in range(self.k):
ids = rng.permutation(X.shape[0])
maybe_inliers = ids[: self.n]
maybe_model = copy(self.model).fit(X[maybe_inliers], y[maybe_inliers])
thresholded = (
self.loss(y[ids][self.n :], maybe_model.predict(X[ids][self.n :]))
< self.t
)
inlier_ids = ids[self.n :][np.flatnonzero(thresholded).flatten()]
if inlier_ids.size > self.d:
inlier_points = np.hstack([maybe_inliers, inlier_ids])
better_model = copy(self.model).fit(X[inlier_points], y[inlier_points])
this_error = self.metric(
y[inlier_points], better_model.predict(X[inlier_points])
)
if this_error < self.best_error:
self.best_error = this_error
self.best_fit = maybe_model
return self
def predict(self, X):
return self.best_fit.predict(X)
def square_error_loss(y_true, y_pred):
return (y_true - y_pred) ** 2
def mean_square_error(y_true, y_pred):
return np.sum(square_error_loss(y_true, y_pred)) / y_true.shape[0]
class LinearRegressor:
def __init__(self):
self.params = None
def fit(self, X: np.ndarray, y: np.ndarray):
r, _ = X.shape
X = np.hstack([np.ones((r, 1)), X])
self.params = np.linalg.inv(X.T @ X) @ X.T @ y
return self
def predict(self, X: np.ndarray):
r, _ = X.shape
X = np.hstack([np.ones((r, 1)), X])
return X @ self.params
if __name__ == "__main__":
regressor = RANSAC(model=LinearRegressor(), loss=square_error_loss, metric=mean_square_error)
X = np.array([-0.848,-0.800,-0.704,-0.632,-0.488,-0.472,-0.368,-0.336,-0.280,-0.200,-0.00800,-0.0840,0.0240,0.100,0.124,0.148,0.232,0.236,0.324,0.356,0.368,0.440,0.512,0.548,0.660,0.640,0.712,0.752,0.776,0.880,0.920,0.944,-0.108,-0.168,-0.720,-0.784,-0.224,-0.604,-0.740,-0.0440,0.388,-0.0200,0.752,0.416,-0.0800,-0.348,0.988,0.776,0.680,0.880,-0.816,-0.424,-0.932,0.272,-0.556,-0.568,-0.600,-0.716,-0.796,-0.880,-0.972,-0.916,0.816,0.892,0.956,0.980,0.988,0.992,0.00400]).reshape(-1,1)
y = np.array([-0.917,-0.833,-0.801,-0.665,-0.605,-0.545,-0.509,-0.433,-0.397,-0.281,-0.205,-0.169,-0.0531,-0.0651,0.0349,0.0829,0.0589,0.175,0.179,0.191,0.259,0.287,0.359,0.395,0.483,0.539,0.543,0.603,0.667,0.679,0.751,0.803,-0.265,-0.341,0.111,-0.113,0.547,0.791,0.551,0.347,0.975,0.943,-0.249,-0.769,-0.625,-0.861,-0.749,-0.945,-0.493,0.163,-0.469,0.0669,0.891,0.623,-0.609,-0.677,-0.721,-0.745,-0.885,-0.897,-0.969,-0.949,0.707,0.783,0.859,0.979,0.811,0.891,-0.137]).reshape(-1,1)
regressor.fit(X, y)
import matplotlib.pyplot as plt
plt.style.use("seaborn-darkgrid")
fig, ax = plt.subplots(1, 1)
# 支持中文
plt.rcParams['font.sans-serif'] = ['SimHei'] # 用来正常显示中文标签
plt.rcParams['axes.unicode_minus'] = False # 用来正常显示负号
plt.scatter(X, y)
plt.title("RANSAC拟合直线")
line = np.linspace(-1, 1, num=100).reshape(-1, 1)
plt.plot(line, regressor.predict(line), c="peru")
plt.show()
RANSAC算法迭代参数的自适应
上面提到的RANSAC算法总要迭代足够的次数才能终止,这就留给设计者一个难题,迭代次数少了,找不到最优解,迭代次数多了,程序耗时增加。接下来给读者介绍一种自适应的迭代次数的方法。
当误差点比例较小时,应尽快停止迭代;否则,算法需要在足够多的迭代后终止,以获得更可靠的结果。
令:RANSAC 算法在运行后提供至少一个有用结果的期望概率。
让满足模型的点/所有检测到的点
令满足当前模型的点数
那么
即
代码实现流程如下
// 以下代码来自OpenCV
/**
* @Method: 更新ransac的迭代次数
* @Returns: int 更新后的最大迭代次数
* @Qualifier:
* @Parameter: double p 信心分数
* @Parameter: double ep 错误点所占的比例
* @Parameter: int modelPoints 模型中点的数量
* @Parameter: int maxIters 当前最大迭代次数
*/
int RANSACUpdateNumIters( double p, double ep, int modelPoints, int maxIters )
{
if( modelPoints <= 0 )
CV_Error( Error::StsOutOfRange, "the number of model points should be positive" );
p = MAX(p, 0.);
p = MIN(p, 1.);
ep = MAX(ep, 0.);
ep = MIN(ep, 1.);
// avoid inf's & nan's
double num = MAX(1. - p, DBL_MIN);
double denom = 1. - std::pow(1. - ep, modelPoints);
if( denom < DBL_MIN )
return 0;
num = std::log(num);
denom = std::log(denom);
return denom >= 0 || -num >= maxIters*(-denom) ? maxIters : cvRound(num/denom);
}
文章出处登录后可见!
已经登录?立即刷新
