参考:
https://github.com/facebookresearch/esm
https://huggingface.co/facebook/esm2_t33_650M_UR50D
https://esmatlas.com/resources?action=fold
1、transformers版本使用
直接输入Fasta 氨基酸序列格式就行;第一次下载esm2_t33_650M_UR50D模型有点慢,有2个多G大
from transformers import BertTokenizer, AutoTokenizer,BertModel, AutoModel
tokenizer_ = AutoTokenizer.from_pretrained("facebook/esm2_t33_650M_UR50D")
model_ = AutoModel.from_pretrained("facebook/esm2_t33_650M_UR50D")
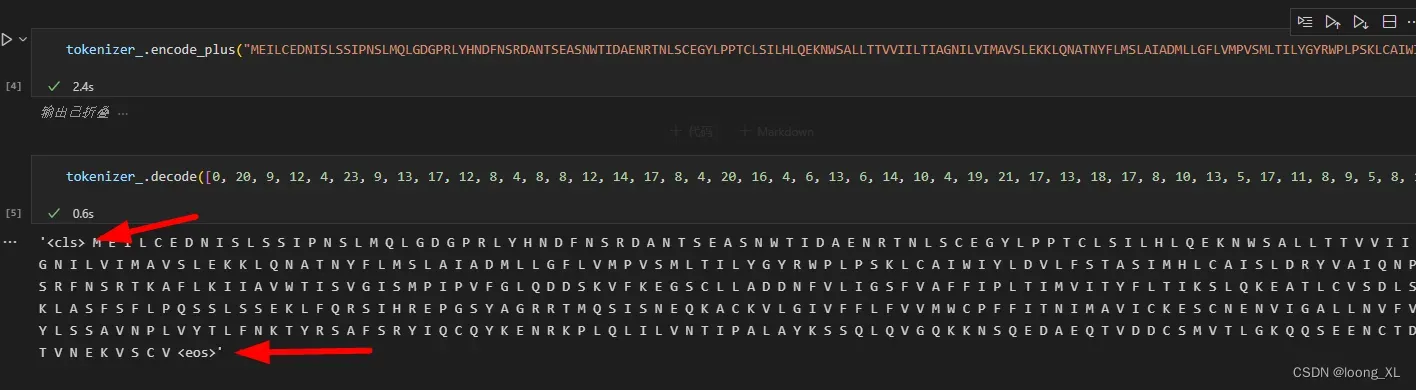
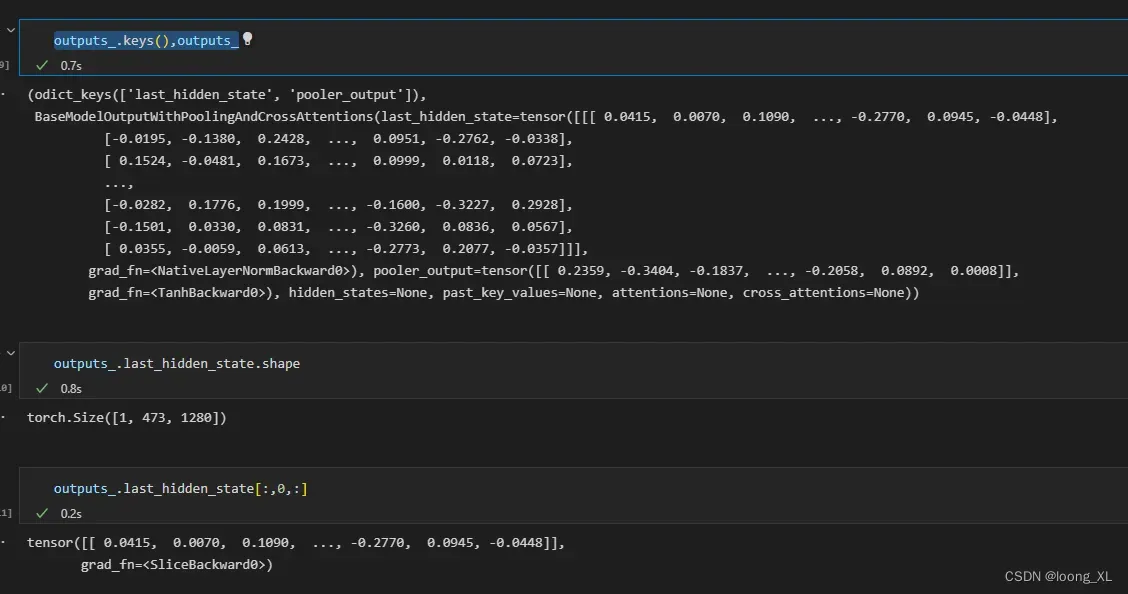
outputs_ = model_(**tokenizer_("MEILCEDNISLSSIPNSLMQLGDGPRLYHNDFNSRDANTSEASNWTIDAENRTNLSCEGYLPPTCLSILHLQEKNWSALLTTVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYRWPLPSKLCAIWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPIPVFGLQDDSKVFKEGSCLLADDNFVLIGSFVAFFIPLTIMVITYFLTIKSLQKEATLCVSDLSTRAKLASFSFLPQSSLSSEKLFQRSIHREPGSYAGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCNENVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENRKPLQLILVNTIPALAYKSSQLQVGQKKNSQEDAEQTVDDCSMVTLGKQQSEENCTDNIETVNEKVSCV", return_tensors='pt'))
outputs_.keys(),outputs_
outputs_.last_hidden_state[:,0,:] ##这是获取得到的 cls 蛋白整体代表的向量
tokenizer_对输入蛋白会头尾添加cls、eos特殊字符,占两个字符长度
2、下载代码离线运行向量表示
参考:https://github.com/facebookresearch/esm
直接clone下来代码
###下载模型
import torch
model, alphabet = torch.hub.load("facebookresearch/esm:main", "esm2_t33_650M_UR50D")
import torch
import esm
# Load ESM-2 model
model, alphabet = esm.pretrained.esm2_t33_650M_UR50D()
batch_converter = alphabet.get_batch_converter()
model.eval() # disables dropout for deterministic results
# Prepare data (first 2 sequences from ESMStructuralSplitDataset superfamily / 4)
data = [
("protein1", "MKTVRQERLKSIVRILERSKEPVSGAQLAEELSVSRQVIVQDIAYLRSLGYNIVATPRGYVLAGG"),
("protein2", "KALTARQQEVFDLIRDHISQTGMPPTRAEIAQRLGFRSPNAAEEHLKALARKGVIEIVSGASRGIRLLQEE"),
("protein2 with mask","KALTARQQEVFDLIRD<mask>ISQTGMPPTRAEIAQRLGFRSPNAAEEHLKALARKGVIEIVSGASRGIRLLQEE"),
("protein3", "K A <mask> I S Q"),
]
batch_labels, batch_strs, batch_tokens = batch_converter(data)
# Extract per-residue representations (on CPU) ## 计算出每个氨基酸的向量,维度1024
with torch.no_grad():
results = model(batch_tokens, repr_layers=[33], return_contacts=True)
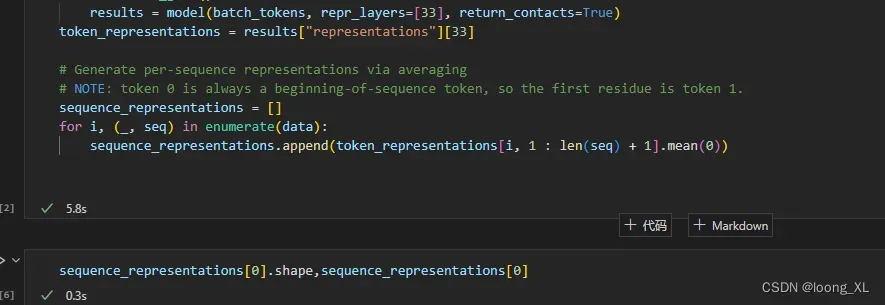
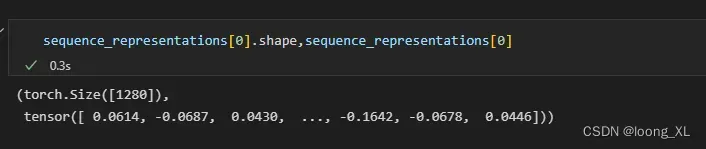
token_representations = results["representations"][33]
# Generate per-sequence representations via averaging ##蛋白质整个的向量,用所有氨基酸的向量平均,维度1024
# NOTE: token 0 is always a beginning-of-sequence token, so the first residue is token 1.
sequence_representations = []
for i, (_, seq) in enumerate(data):
sequence_representations.append(token_representations[i, 1 : len(seq) + 1].mean(0))
# Look at the unsupervised self-attention map contact predictions
import matplotlib.pyplot as plt
for (_, seq), attention_contacts in zip(data, results["contacts"]):
plt.matshow(attention_contacts[: len(seq), : len(seq)])
plt.title(seq)
plt.show()
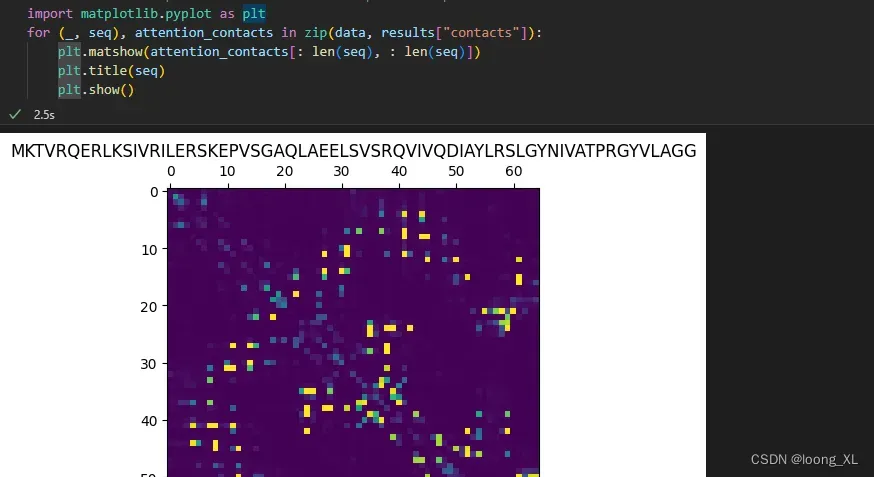
单个蛋白
#Prepare data (first 2 sequences from ESMStructuralSplitDataset superfamily / 4)
data = [
("protein111", "MEILCEDNISLSSIPNSLMQLGDGPRLYHNDFNSRDANTSEASNWTIDAENRTNLSCEGYLPPTCLSILHLQEKNWSALLTTVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYRWPLPSKLCAIWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPIPVFGLQDDSKVFKEGSCLLADDNFVLIGSFVAFFIPLTIMVITYFLTIKSLQKEATLCVSDLSTRAKLASFSFLPQSSLSSEKLFQRSIHREPGSYAGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCNENVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENRKPLQLILVNTIPALAYKSSQLQVGQKKNSQEDAEQTVDDCSMVTLGKQQSEENCTDNIETVNEKVSCV"),
]
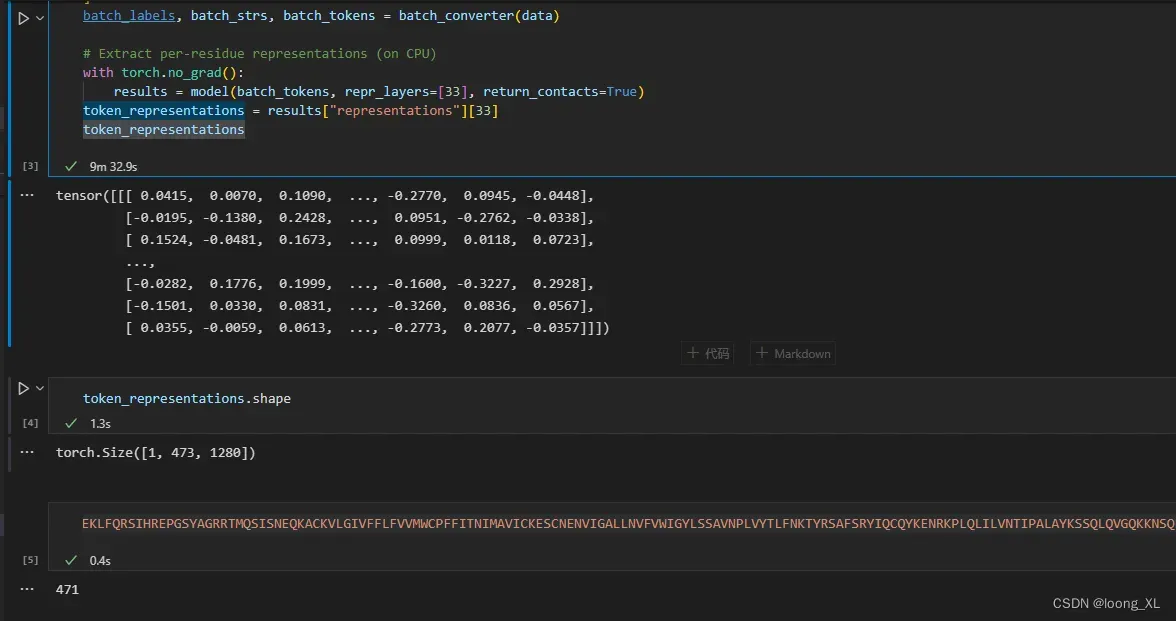
batch_labels, batch_strs, batch_tokens = batch_converter(data)
# Extract per-residue representations (on CPU)
with torch.no_grad():
results = model(batch_tokens, repr_layers=[33], return_contacts=True)
token_representations = results["representations"][33]
token_representations
文章出处登录后可见!
已经登录?立即刷新
